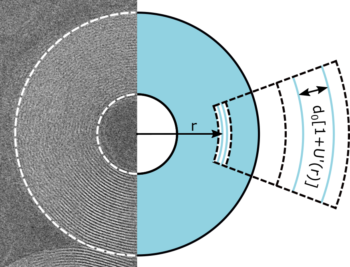
DNA packaging within viruses, chromosomes or nano-devices leads to extremely compact assemblies where not only high density but also very high curvature prevail. Yet DNA is a rather stiff filament, which resists bending. In addition, the surface of DNA is negatively charged, and it is necessary to overcome its self-repulsion to obtain compact forms. The microscopic forces at work are still poorly understood. Multivalent cations can induce an attraction between DNA filaments, but our understanding of this attraction to generate dense and highly curved assemblies remains incomplete, partly because available experimental data are obtained with large samples of uncurved DNA helices. In collaboration with Luca Barberi and Martin Lenz at LPTMS, we analyzed the attraction between DNA filaments folded into nanometric toroidal geometries. By combining cryo-electron microscopy with a mathematical model, we demonstrate a link between the attraction force and the geometry. In particular, we showed that the attraction is weaker when the filaments are bent than when they are straight. In addition, the ability of DNA filaments to attract each other depends on their lateral alignment : DNA double helices can be aligned either in register or out of register, and an optimal alignment cannot be obtained in curved assemblies. These results will not only provide a better understanding of the complex process of DNA packaging in viruses and cells, but could help develop robust designs in DNA nanotechnologies.
These results have been published in Nucleic Acids Research doi.org/10.1093/nar/gkab197
Contact : Amélie Leforestier