Bacterial viruses, also called bacteriophages, are complex macromolecular assemblies. Their genome consists of a unique double-stranded DNA chain (tens of μm in length) confined at high density in the volume of the protein icosahedral capsid (typically tens of nm in diameter). In contrast with eukaryotic viruses, only DNA enter the host cell after recognition and fixation on a receptor localised at the surface of the bacteria. Symetrically they package their genome into preformed capsids. On account of its physical characteristics, the DNA molecule, a highly charged semi-flexible poly-anion, is difficult to compress and bend into the small capsid volume. To achieve this process, a motor protein generating forces larger than 50 pN. This unique mechanism results in intracapsid pressures of about 50 atmospheres, necessary to deliver the genome to the next host cell.
Controlling DNA ejection in vitro, upon interaction of the phage with its purified bacterial receptor, allows us : i) to monitor the length of the DNA chain trapped in the capsid, ii) to modify the ionic environment of the molecule (the capsid is permeable to ions and water), and iii) to visualize DNA individual chains inside the capsid using cryo-EM.
We focus on the organization and conformation of the DNA molecule confined and compressed inside the protein capsid. We analyse phase transitions occuring upon decrease of DNA concentration upon ejection of upon changes in the physico-chemical environment (temperature, osmotic pressure, ionic environment).
Thanks to this versatile system we also explore the behaviour of confined single DNA chains under controlled conditions and question the relationship between genome transfer and organisation.
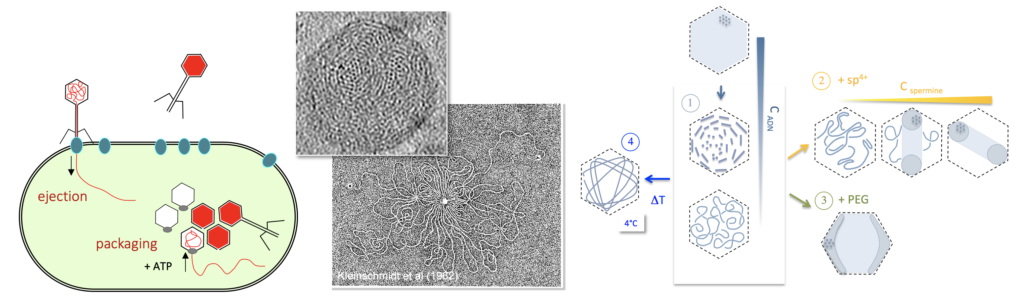
From left to right: sketch of the bacteriophage infection cycle; DNA molecule highly compacted within the capsid; polymorphism of DNA organisation in phage capsids as a function of DNA concentration, temperature presence/absence of condensig concentrations of multivalent cations.
Related publications
- Leforestier, A., Brasiles, S., de Frutos, M., Raspaud, E., Letellier, L., Tavares, P., & Livolant, F. (2008). Bacteriophage T5 DNA ejection under pressure. Journal of molecular biology, 384(3), 730-739. doi.org/10.1016/j.jmb.2008.09.035
- Leforestier, A., & Livolant, F. (2009). Structure of toroidal DNA collapsed inside the phage capsid. Proceedings of the National Academy of Sciences, 106(23), 9157-9162. doi.org/10.1073/pnas.0901240106
- Leforestier, A., & Livolant, F. (2010). The bacteriophage genome undergoes a succession of intracapsid phase transitions upon DNA ejection. Journal of molecular biology, 396(2), 384-395. doi.org/10.1016/j.jmb.2009.11.047
- Leforestier, A., Šiber, A., Livolant, F., & Podgornik, R. (2011). Protein-DNA interactions determine the shapes of DNA toroids condensed in virus capsids. Biophysical journal, 100(9), 2209-2216. doi.org/10.1016/j.bpj.2011.03.012
- Leforestier, A. (2013). Polymorphism of DNA conformation inside the bacteriophage capsid. Journal of biological physics, 39(2), 201-213. doi.org/10.1007/s10867-013-9315-y
- Sung, B., Leforestier, A., & Livolant, F. (2016). Coexistence of coil and globule domains within a single confined DNA chain. Nucleic acids research, 44(3), 1421-1427. doi.org/10.1093/nar/gkv1494
- Frutos, M. D., Leforestier, A., Degrouard, J., Zambrano, N., Wien, F., Boulanger, P., … & Livolant, F. (2016). Can changes in temperature or ionic conditions modify the DNA organization in the full bacteriophage capsid?. The Journal of Physical Chemistry B, 120(26), 5975-5986. doi.org/10.1021/acs.jpcb.6b01783
- Tresset, G., Castelnovo, M., & Leforestier, A. (2017). Assemblage et désassemblage des virus: mode d’emploi. Reflets de la physique, (52), 22-26. doi.org/10.1051/refdp/201752022
Contact
Amélie Leforestier: amelie.leforestier@universite-paris-saclay.fr