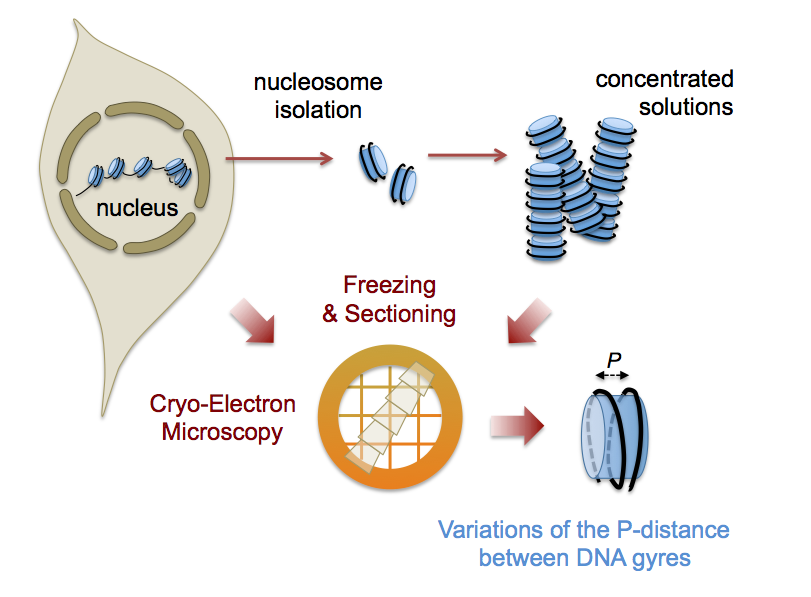
In Eukaryotes, DNA is regularly wound into a left-handed super-helix around the histone octamer to form a chain of nucleosomes. Atomic structures have been obtained by crystallography of engineered nucleosomes. However nucleosomes are now being recognised as pleiomorphic and dynamical entities, with a multiplicity of states related to variability in their composition, their interaction with other molecules, as well as to their dynamics. But nucleosome conformation, while documented in vitro and in silico, is unknown in the cellular context. In collaboration with M. Eltsov (Goethe University, Frankfurt), we explored interphase nuclei of different cell types (human cell lines and Drosophila embryo) using cryo-electron microscopy and tomography of vitreous sections. For the first time, we visualized individual nucleosomes at a level of detail that allows us to retrieve structural information. Measuring the distance between the DNA gyres of the super-helix , we showed that nucleosome conformation is variable and on average more open than the canonical structures. To understand this phenomenon, we used isolated nucleosomes and demonstrated a salt-dependent transition, with a high salt compact conformation resembling the canonical nucleosome and an open low salt one, closer to nuclear nucleosomes.
This work opens a route to chromatin exploration in situ. Further nucleosome characterization and cartography are now needed to question the relationship between this conformational variability and chromatin functional states.
Nucleic Acids Research, Volume 46 (17), 28 September 2018, Pages 9189–9200, doi.org/10.1093/nar/gky670
Contact : Amélie Leforestier