Mohamad HARASTANI has defended his PhD thesis entitled “Image analysis method development for in vitro and in situ cryo electron tomography studies of conformational variability of biomolecular complexes: case of nucleosome structural and dynamics studies”, under the supervision of Slavica JONIC (IMPMC, Sorbonne Université) and Amélie LEFORESTIER.

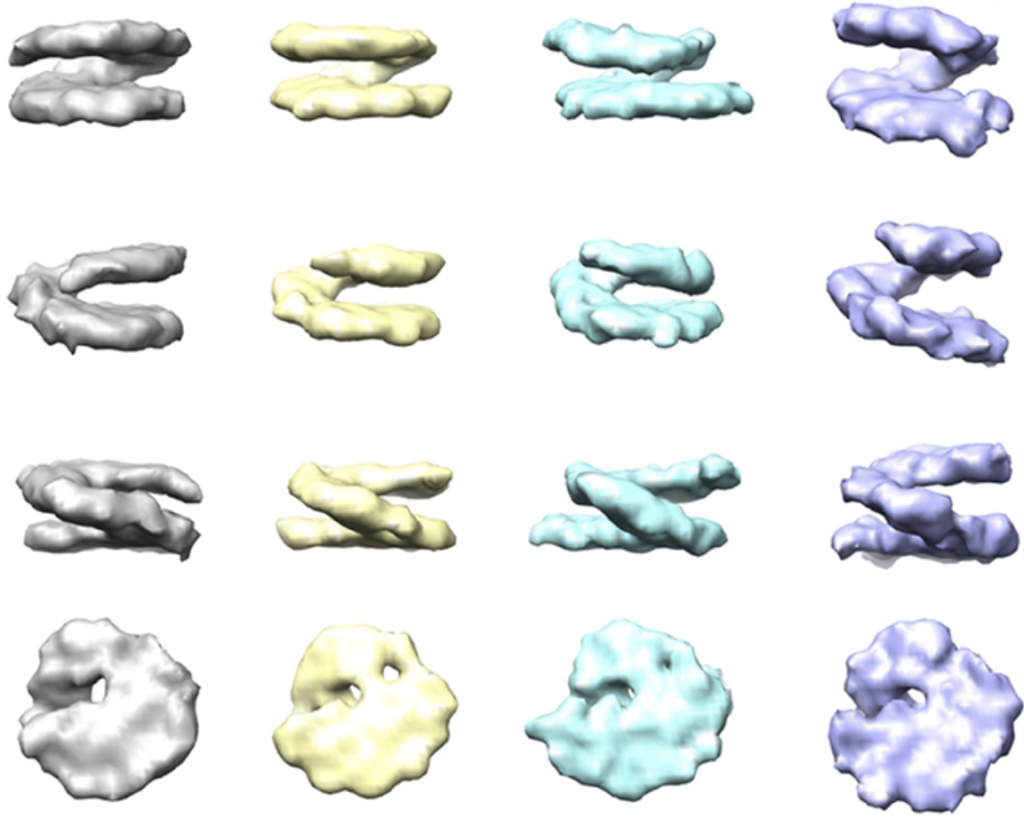
This thesis presents the first two cryo-electron tomography (cryo-ET) data processing methods that address continuous conformational variability of biomolecules, HEMNMA-3D and TomoFlow. HEMNMA-3D analyses experimental data with motion directions simulated by Normal Mode Analysis, and allows the discovery of a large range of biomolecular motions hidden in the data. However, HEMNMA-3D depends on this prior (simulated motion directions), making it prone to misinterpretation and bias when misused. TomoFlow extracts movements from the data without prior information using a computer vision technique called the Optical Flow. Therefore, it is less prone to misinterpretation and misuse. However, when it encounters large motion magnitudes, it results in a smooth and downscaled version of the actual biomolecular motion. HEMNMA-3D and TomoFlow have different mathematical models, but both are able to explore biomolecular conformational landscapes and are superior to classification. HEMNMA-3D and TomoFlow are validated on synthetic datasets. The two methods are applied on experimental cryo-ET data of nucleosome conformational variability, a specially challenging object in situ. They show coherent results, shedding insight into the conformational variability of nucleosomes in situ, in line with preliminary and theoretical analyses. Based on this, they are also expected to be useful for conformational studies of other biomolecular complexes in vitro and in situ. The software of HEMNMA-3D and TomoFlow is publicly available, as part of the cryo-ET data processing pipeline of the open-source software package ContinuousFlex, which is currently a plugin of Scipion software, extensively used in the field, and uses Scipion’s backend software Xmipp.